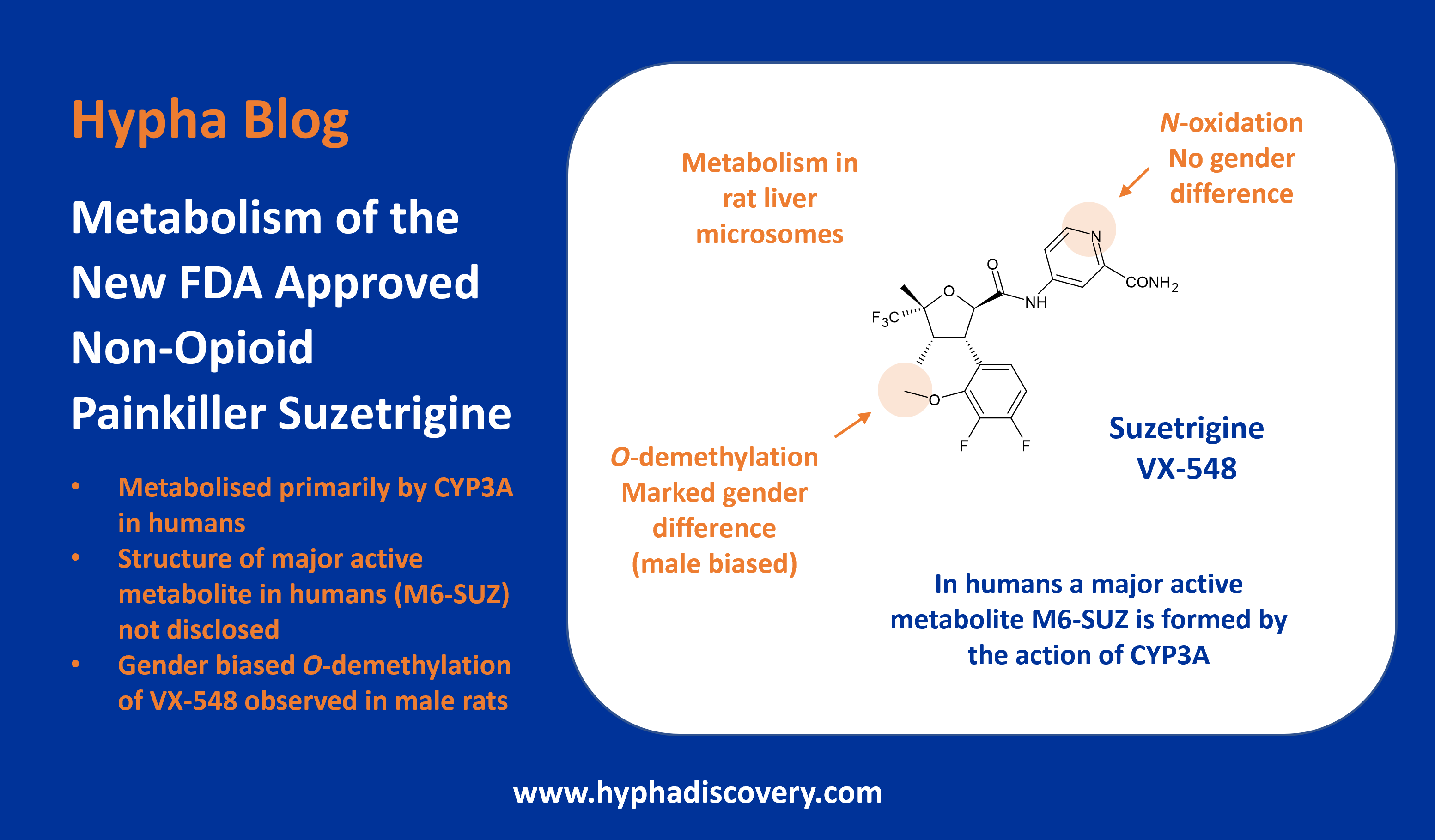
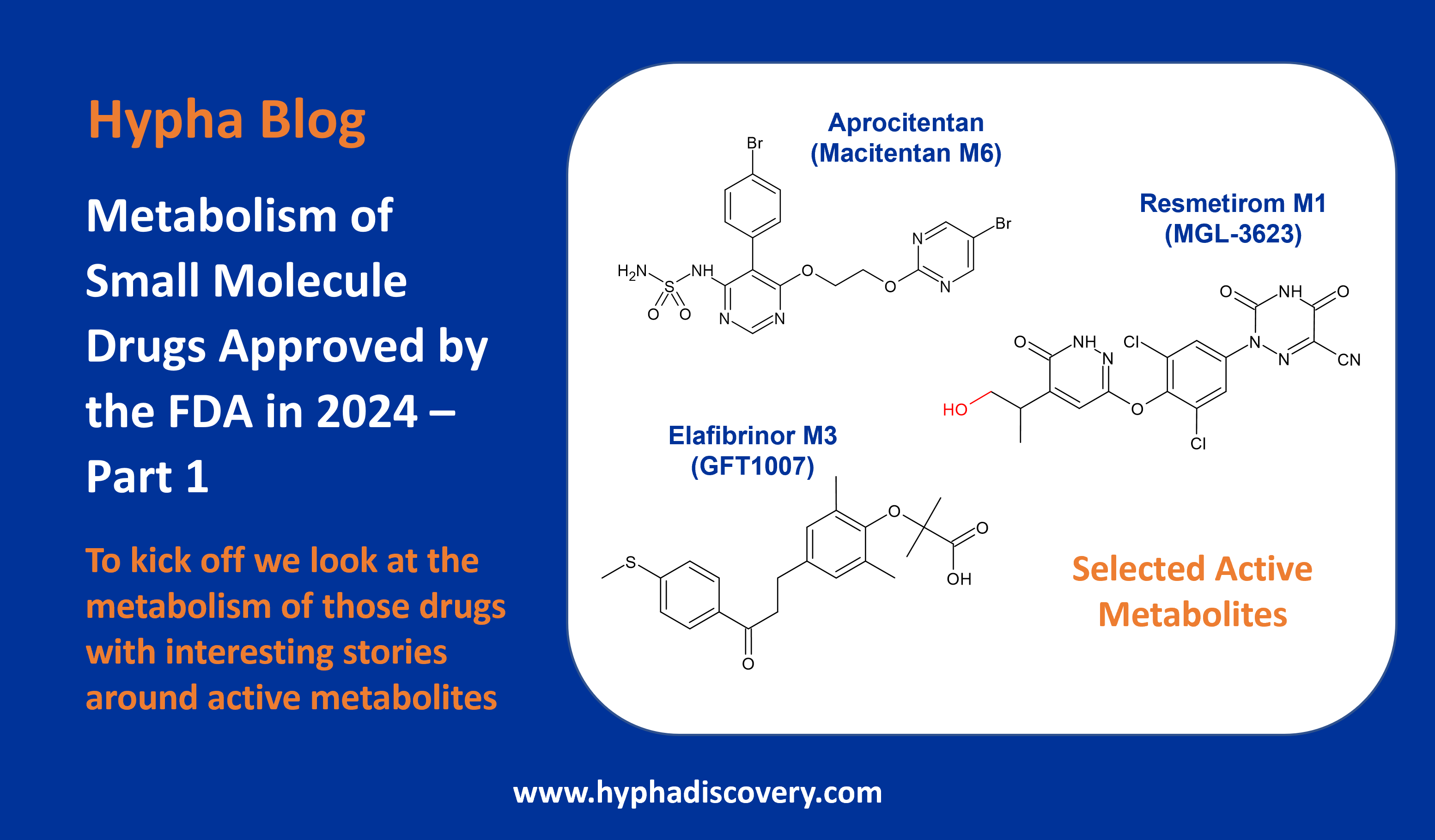
First-ever identification of an aldehyde oxidase-mediated DDI
Inhibition of aldehyde oxidase mediated metabolism of OS1-930 by erlotinib
Where metabolic pathways have not been interrogated for drugs in development, unwelcome surprises may arise. Scientists at Pfizer recently uncovered a major role for aldehyde oxidase in the metabolism of quinoline-containing OSI-930, and demonstrated an aldehyde oxidase-mediated DDI (drug-drug interaction) arising from inhibition of the enzyme by erlotinib (Tang et al., 2024).
OSI-930 is an inhibitor of c-Kit and vascular endothelial growth factor receptor 2. Aspects of its metabolism have previously been reported in the literature but with no mention of the involvement of aldehyde oxidase (AO). In this latest study by Tang et al. using human hepatocytes, AO is observed as a major route of metabolism, resulting in a mono-oxygenated 2-oxo metabolite (M460c/M1). Reaction phenotyping experiments in human hepatocytes suggest that AO contributes almost 50% to the clearance of OSI-930.
Non-AO mediated metabolism resulted in two other mono-oxygenated metabolites, M460a and M460b. M460a results from oxidation of the thiophene ring from which several conjugates were observed. It is known that cytochrome P450s (CYPs) catalyse the oxidation of thiophenes with the simultaneous formation of two reactive intermediates, a thiophene-S-oxide and a thiophene epoxide (Dansette et al., 2005). The presence of M460a and related metabolites correlates with the previously reported oxidation of OSI-930 by CYPs to an apparent sulfoxide which reacts via Michael-addition to the 5-position of the thiophene ring and the formation of glutathione conjugates (Medower et al., 2008). Additionally, oxidation of the thiophene ring to an epoxide is possible to form a dihydrodiol which rearomatises to phenol and aromatic GSH adducts. M460b is formed by likely CYP-mediated hydroxylation of the methylene group at the 4-position of the quinoline ring.
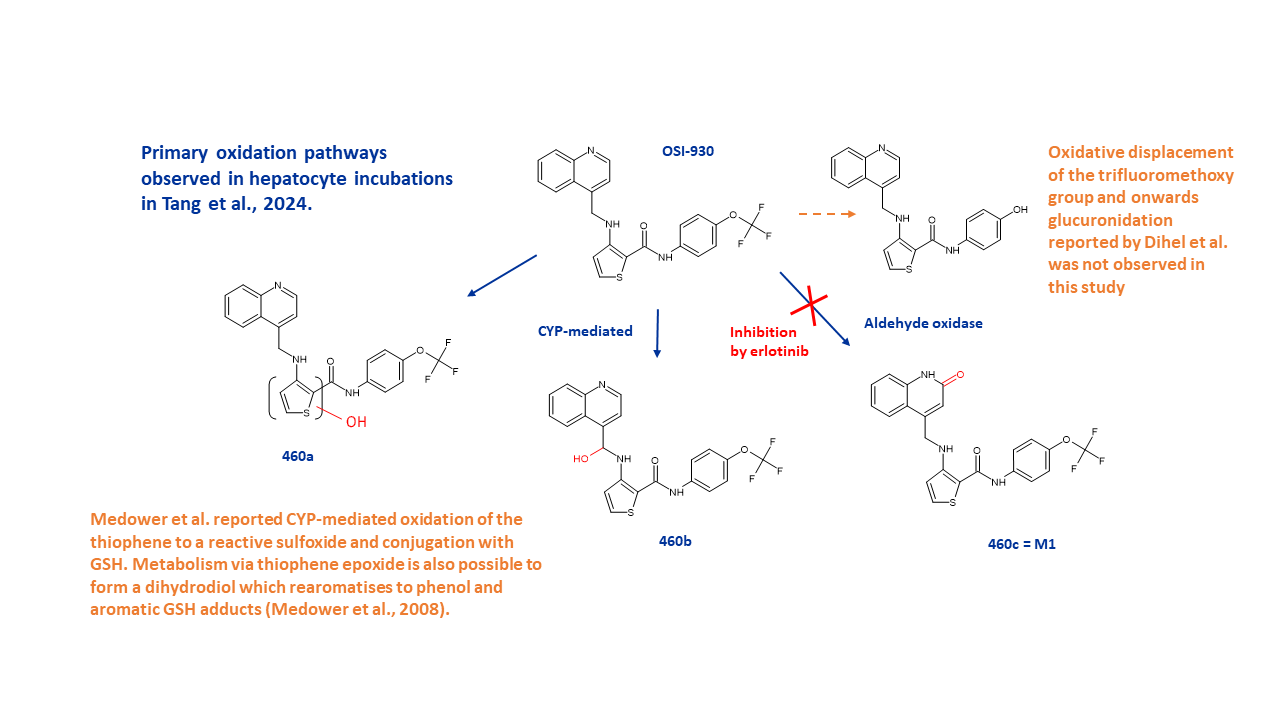
Primary oxidation pathways observed in hepatocyte incubations in Tang et al., 2024.
Not observed was a previously reported metabolite arising from oxidative displacement of the trifluoromethoxy group via ipso-substitution which results in displacement of aromatic ring substituents. In vivo the resulting hydroxylated metabolite is then glucuronidated (Dihel et al., 2009).
Several competitive inhibitors of AO have been reported, including the epidermal growth factor receptor tyrosine kinase inhibitors erlotinib, gefitinib and their metabolites. These compounds possess a quinazoline moiety that fits well into the active site of the enzyme. Erlotinib was found to be a particularly potent inhibitor (Tan et al., 2020).
Use of a mechanistic static model by Tang and colleagues predicted a ~1.85-fold increase in the systemic exposure of OSI-930 caused by inhibition of the AO route of metabolism by erlotinib, supporting previously reported clinical observations and the first-ever identification of an AO-mediated DDI.
References
Dansette PM, Bertho G, Mansuy D. First evidence that cytochrome P450 may catalyze both S-oxidation and epoxidation of thiophene derivatives. Biochem Biophys Res Commun. 2005; 338(1):450-455. https://doi.org/10.1016/j.bbrc.2005.08.091
Dihel L, Kittleson C, Mulvihill K, Johnson WW. Oxidative metabolism of the trifluoromethoxy moiety of OSI-930. Drug Metabol Drug Interact. 2009;24(2-4):95-121. https://doi.org/10.1515/DMDI.2009.24.2-4.95
Medower C, Wen L, and Johnson WW. Cytochrome P450 Oxidation of the Thiophene Containing Anticancer Drug 3-[(Quinolin-4-ylmethyl)-amino]-thiophene-2-carboxylic Acid (4-Trifluoromethoxy-phenyl)-amide to an Electrophilic Intermediate. Chem. Res. Toxicol. 2008, 21, 8, 1570–1577. https://doi.org/10.1021/tx700430n
Wee Kiat Tan, Alyssa Rui Yi Tan, Punitha Sivanandam, Ernest Jing Hui Goh, Ze Ping Yap, Nur Fazilah Saburulla, Karl Austin-Muttitt, Jonathan G.L. Mullins and Aik Jiang Lau.In Vitro Inhibition of Human Aldehyde Oxidase Activity by Clinically Relevant Concentrations of Gefitinib and Erlotinib: Comparison with Select Metabolites, Molecular Docking Analysis, and Impact on Hepatic Metabolism of Zaleplon and Methotrexate. Journal of Pharmacology and Experimental Therapeutics August 1, 2020, 374 (2) 295-307; DOI: https://doi.org/10.1124/jpet.120.265249
Lloyd Wei Tat Tang, Yuanyuan Shi, Raman Sharma and R. Scott Obach. AO-mediated DDI between Erlotinib and OSI-930. Drug Metabolism and Disposition June 18, 2024, DMD-AR-2024-001802; DOI: https://doi.org/10.1124/dmd.124.001802